Implementing PCA from Scratch
Principal Component Analysis in Python and NumPy
Last update: 26.03.2023. All opinions are my own.
1. Overview
This blog post provides a tutorial on implementing the Principal Component Analysis algorithm using Python and NumPy. We will set up a simple class object, implement relevant methods to perform the decomposition, and illustrate how it works on a toy dataset.
Why are we implementing PCA from scratch if the algorithm is already available in scikit-learn? First, coding something from scratch is the best way to understand it. You may know many ML algorithms, but being able to write it down indicates that you have really mastered it. Second, implementing algorithms from scratch is a common task in ML interviews in tech companies, which makes it a useful skill that a job candidate should practice. Last but not least, it's a fun exercise, right? :)
This post is part of "ML from Scratch" series, where we implement established ML algorithms in Python. Check out other posts to see further implementations.
2. How PCA works
Before jumping to implementation, let's quickly refresh our minds. How does PCA work?
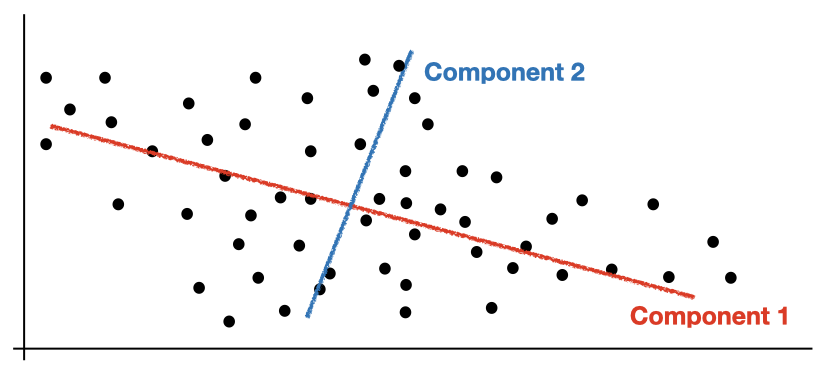
PCA is a popular unsupervised algorithm used for dimensionality reduction. In a nutshell, PCA helps you to reduce the number of feature in your dataset by combining the features without loosing too much information. More specifically, PCA finds a linear data transformation that projects the data into a new coordinate system with a fewer dimensions. To capture the most variation in the original data, this projection is done by finding the so-called principal components - eigenvectors of the data's covariance matrix - and multiplying the actual data matrix with a subset of the components. This procedure is what we are going to implement.
P.S. If you need a more detailed summary of how PCA works, check out this Wiki page.
import numpy as np
In line with object-oriented programming practices, we will implement PCA as a class with a set of methods. We will need the following five:
-
__init__(): initialize the class object. -
fit(): center the data and identify principal components. -
transform(): transform new data into the identified components.
Let's sketch a class object template. Since we implement functions as class methods, we include self argument for each method:
class PCA:
def __init__(self):
pass
def fit(self):
"""
Find principal components
"""
pass
def transform(self):
"""
Transform new data
"""
pass
Now let's go through each method one by one.
The __init__() method is run once when the initialize the PCA class object.
One thing need to do on the initialization step is to store meta-parameters of our algorithm. For PCA, there is only one meta-parameter we will specify: the number of components. We will save it as self.num_components.
Apart from the meta-parameters, we will create three placeholders that we will use to store important class attributes:
-
self.components: array with the principal component weights -
self.mean: mean variable values observed in the training data -
self.variance_share: proportion of variance explained by principal components
def __init__(self, num_components):
self.num_components = num_components
self.components = None
self.mean = None
self.variance_share = None
Next, let's implement the fit() method - the heart of our PCA class. This method will be applied to a provided dataset to identify and memorize principal components.
We will do the following steps:
- Center the data by subtracting the mean values for each variable. Normalizing variables is important to make sure that their impact in the data variation is similar and does not depend on the range of that variable. We will also memorize the mean values as
self.meanas we will need it later for the data transformation. - Calculate eigenvectors of the covariance matrix. First, we will use
np.cov()to get the covariance matrix of the data. Next, we will leveragenp.linalg.eig()to do the eigenvalue decomposition and obtain both eigenvalues and eigenvectors. - Sort eigenvalues and eigenvectors in the decreasing order. Since we will use a smaller number of components compared to the number of variables in the original data, we would like to focus on components that reflect more data variation. In our case, eigenvectors that correspond to larger eigenvalues capture more variation.
- Store an array with the top
num_componentscomponents asself.components.
Finally, we will calculate and memorize the data variation explained by the selected components as self.variance_share. This can be computed as a cumulative sum of the corresponding eigenvalues divided by the total sum of eigenvalues.
def fit(self, X):
"""
Find principal components
"""
# data centering
self.mean = np.mean(X, axis = 0)
X -= self.mean
# calculate eigenvalues & vectors
cov_matrix = np.cov(X.T)
values, vectors = np.linalg.eig(cov_matrix)
# sort eigenvalues & vectors
sort_idx = np.argsort(values)[::-1]
values = values[sort_idx]
vectors = vectors[:, sort_idx]
# store principal components & variance
self.components = vectors[:self.num_components]
self.variance_share = np.sum(values[:self.num_components]) / np.sum(values)
The most difficult part is over! Last but not least, we will implement a method to perform the data transformation.
This will be run after calling the fit() method on the training data, so we only need to implement two steps:
- Center the new data using the same mean values that we used on the fitting stage.
- Multiply the data matrix with the matrix of the selected components. Note that we will need to transpose the components matrix to ensure the right dimensionality.
def transform(self, X):
"""
Transform data
"""
# data centering
X -= self.mean
# decomposition
return np.dot(X, self.components.T)
Putting everything together, this is how our implementation looks like:
class PCA:
def __init__(self, num_components):
self.num_components = num_components
self.components = None
self.mean = None
self.variance_share = None
def fit(self, X):
"""
Find principal components
"""
# data centering
self.mean = np.mean(X, axis = 0)
X -= self.mean
# calculate eigenvalues & vectors
cov_matrix = np.cov(X.T)
values, vectors = np.linalg.eig(cov_matrix)
# sort eigenvalues & vectors
sort_idx = np.argsort(values)[::-1]
values = values[sort_idx]
vectors = vectors[:, sort_idx]
# store principal components & variance
self.components = vectors[:self.num_components]
self.variance_share = np.sum(values[:self.num_components]) / np.sum(values)
def transform(self, X):
"""
Transform data
"""
# data centering
X -= self.mean
# decomposition
return np.dot(X, self.components.T)
X_old = np.random.normal(loc = 0, scale = 1, size = (1000, 10))
X_new = np.random.normal(loc = 0, scale = 1, size = (500, 10))
print(X_old.shape, X_new.shape)
Now, let's instantiate our PCA class, fit it on the old data and transform both datasets!
To see if the algorithm works properly, we will generate four new examples as X_new, gradually increasing the feature values from 1 to 5. We expect the label predicted by KNN to increase from 0 to 1, since we are getting closer to examples in X1. Let's check!
# initialize PCA object
pca = PCA(num_components = 8)
# fit PCA on old data
pca.fit(X_old)
# check explained variance
print(f"Explained variance: {pca.variance_share:.4f}")
Eight components explain more than 83% of the data variation. Not bad! Let's transform the data:
# transform datasets
X_old = pca.transform(X_old)
X_new = pca.transform(X_new)
print(X_old.shape, X_new.shape)
Yay! Everything works as expected. The new datasets have eight features instead of the original ten features.
5. Closing words
This is it! I hope this tutorial helps you to refresh your memory on how PCA works and gives you a good idea on how to implement it yourself. You are now well-equipped to do this exercise on your own!
If you liked this tutorial, feel free to share it on social media and buy me a coffee :) Don't forget to check out other posts in the "ML from Scratch" series. Happy learning!
Liked the post? Share it on social media!
You can also buy me a cup of coffee to support my work. Thanks!
